|
RepeatMasker Open-3-1.6 Release Notes |

|
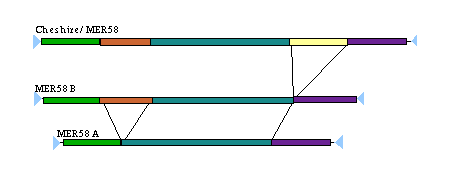 The figure at the right provides an example of a related DNA Transposon family.
Deletion products such as MER58b and MER58a are common among DNA Transposons and create a challenge for classifying
subfragments of an element. For many years RepeatMasker has used a mechanism whereby deletion product fragments
were translated to parent element coordinates prior to grouping annotations. This works well while the relationships
among the elements is well understood and consistent.
The figure at the right provides an example of a related DNA Transposon family.
Deletion products such as MER58b and MER58a are common among DNA Transposons and create a challenge for classifying
subfragments of an element. For many years RepeatMasker has used a mechanism whereby deletion product fragments
were translated to parent element coordinates prior to grouping annotations. This works well while the relationships
among the elements is well understood and consistent.As the repeat databases have grown so has the complexity of relationships between repeat families. In this version of RepeatMasker we have developed a new method for classifying and grouping related transposon fragments. The new method utilizes a set of auto-generated family-family similarity alignments to identify ambiguous fragments and their possible family associations. A new option in RepeatMasker ( -lcambig ) will identify these ambiguous fragments in the .out file with lowercase characters. All non-ambiguous fragments are labeled in uppercase. The following example illustrates how various fragments of the MER58/Cheshire subfamilies would be annotated.
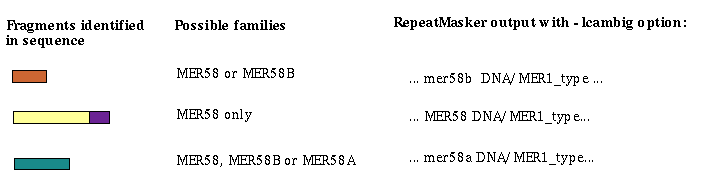
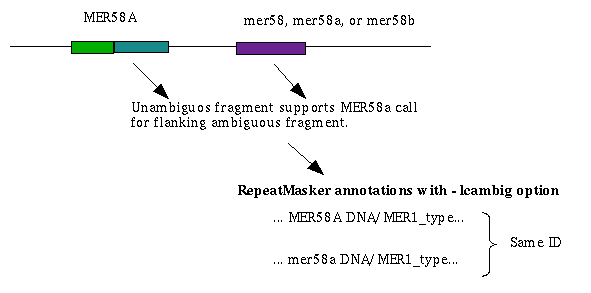
Ambiguous elements may have 3 or more possible family classifications. The one chosen is based on supporting evidence
from flanking annotations. In some cases there is one related non-ambiguous element which, when combined with this
element uniquely identifies which family is the correct choice. For example: